Completed first draft of evaluation section
Showing
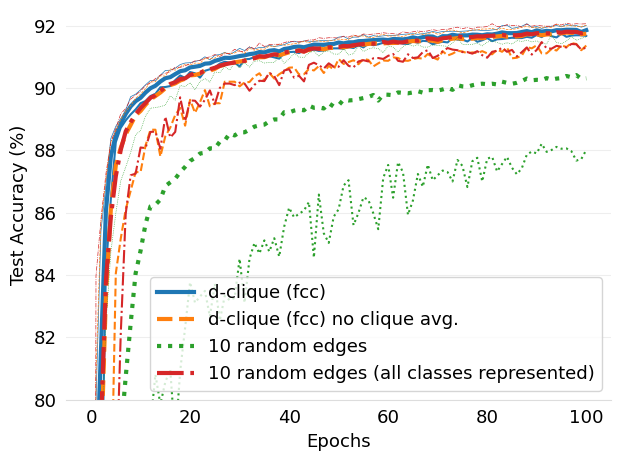
- figures/d-cliques-mnist-linear-comparison-to-non-clustered-topologies.png 0 additions, 0 deletions...s-mnist-linear-comparison-to-non-clustered-topologies.png
- mlsys2022style/d-cliques.tex 2 additions, 1 deletionmlsys2022style/d-cliques.tex
- mlsys2022style/exp.tex 219 additions, 145 deletionsmlsys2022style/exp.tex
- mlsys2022style/figures/convergence-cifar10-random-vs-d-cliques-2-shards.png 0 additions, 0 deletions...ures/convergence-cifar10-random-vs-d-cliques-2-shards.png
- mlsys2022style/figures/convergence-mnist-random-vs-d-cliques-2-shards.png 0 additions, 0 deletions...igures/convergence-mnist-random-vs-d-cliques-2-shards.png
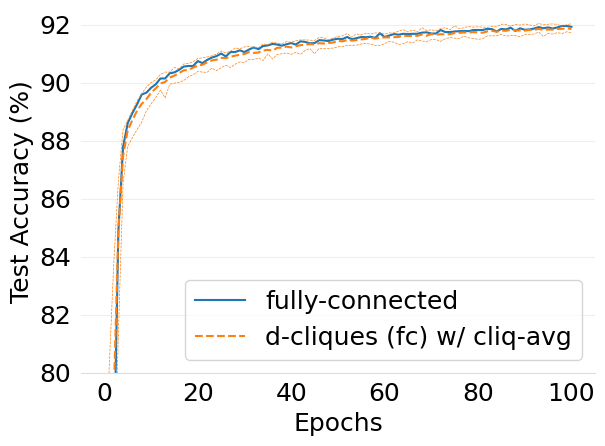
- mlsys2022style/figures/convergence-speed-cifar10-dc-fc-vs-fc-2-shards-per-node.png 0 additions, 0 deletions...nvergence-speed-cifar10-dc-fc-vs-fc-2-shards-per-node.png
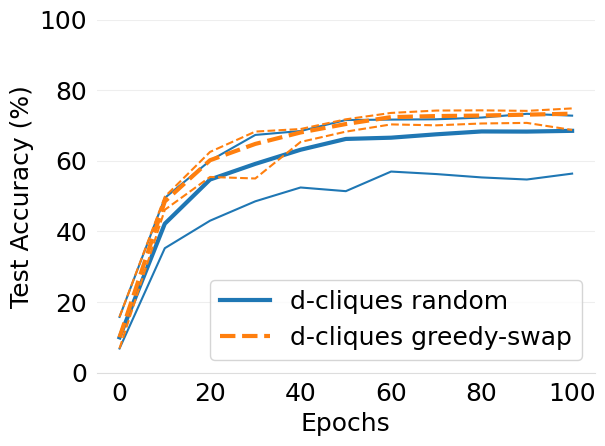
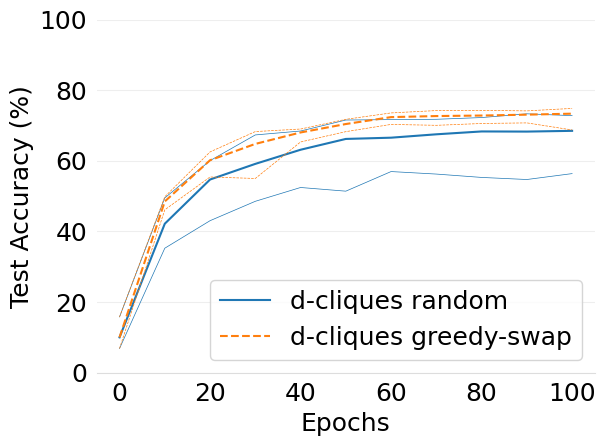
- mlsys2022style/figures/convergence-speed-cifar10-dc-random-vs-dc-gs-2-shards-per-node.png 0 additions, 0 deletions...ce-speed-cifar10-dc-random-vs-dc-gs-2-shards-per-node.png
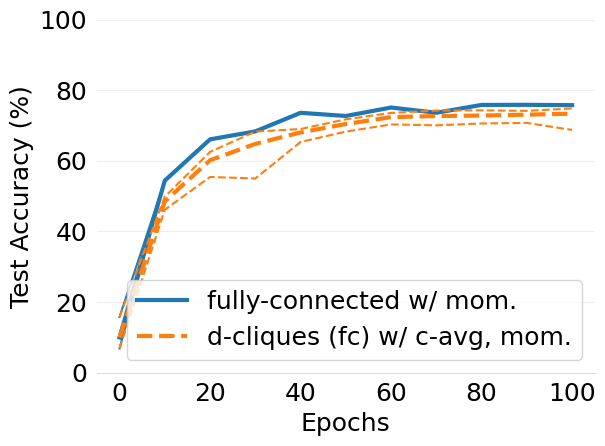
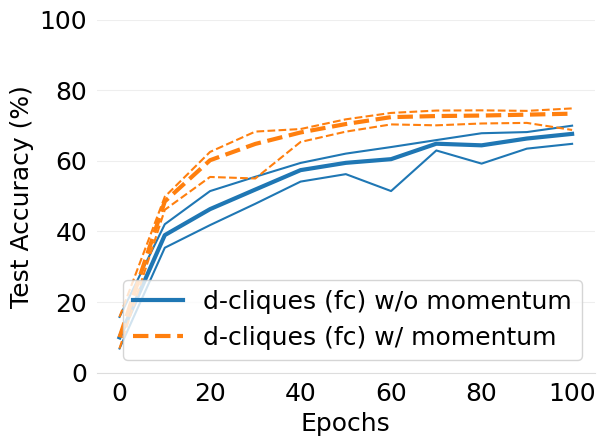
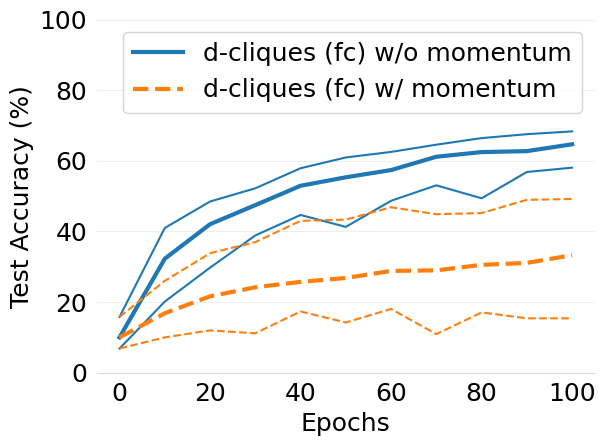
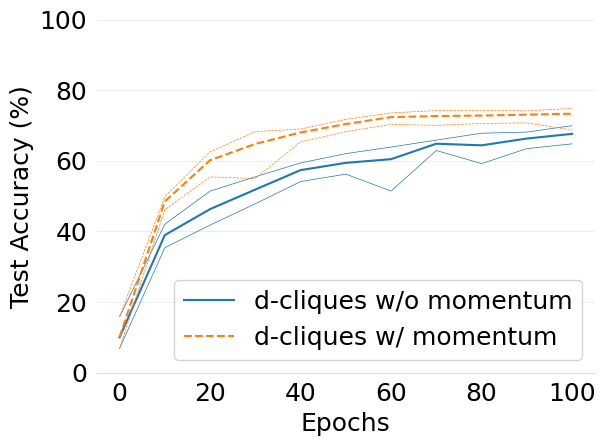
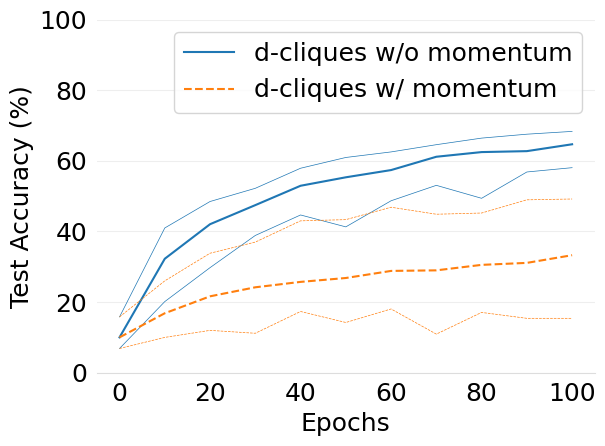
- mlsys2022style/figures/convergence-speed-cifar10-w-c-avg-no-mom-vs-mom-2-shards-per-node.png 0 additions, 0 deletions...speed-cifar10-w-c-avg-no-mom-vs-mom-2-shards-per-node.png
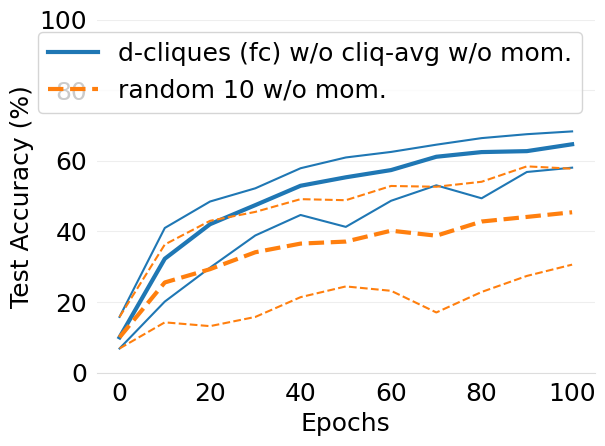
- mlsys2022style/figures/convergence-speed-cifar10-wo-c-avg-no-mom-vs-mom-2-shards-per-node.png 0 additions, 0 deletions...peed-cifar10-wo-c-avg-no-mom-vs-mom-2-shards-per-node.png
- mlsys2022style/figures/convergence-speed-mnist-dc-fc-vs-fc-2-shards-per-node.png 0 additions, 0 deletions...convergence-speed-mnist-dc-fc-vs-fc-2-shards-per-node.png
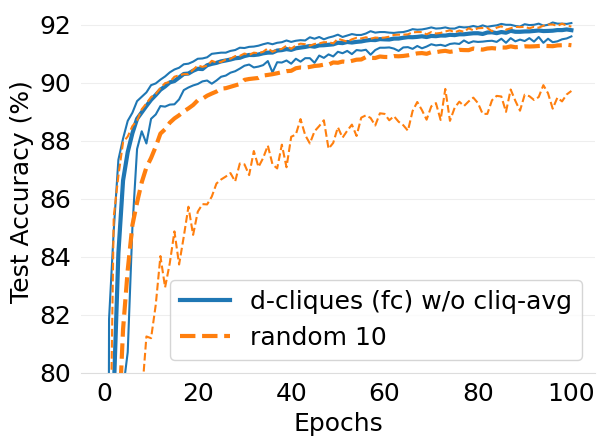
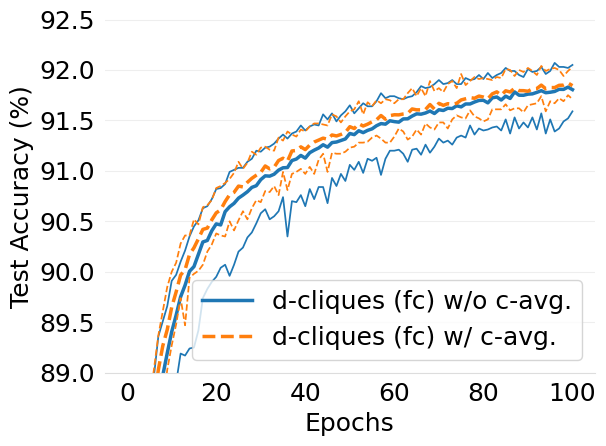
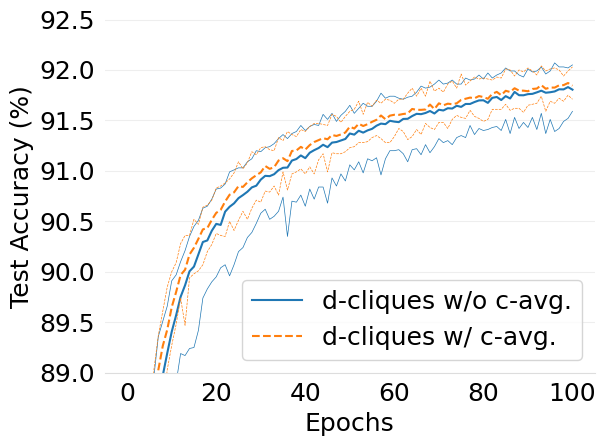
- mlsys2022style/figures/convergence-speed-mnist-dc-no-c-avg-vs-c-avg-2-shards-per-node.png 0 additions, 0 deletions...ce-speed-mnist-dc-no-c-avg-vs-c-avg-2-shards-per-node.png
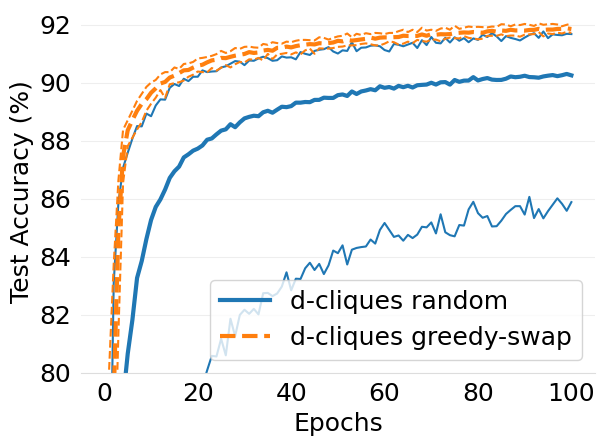
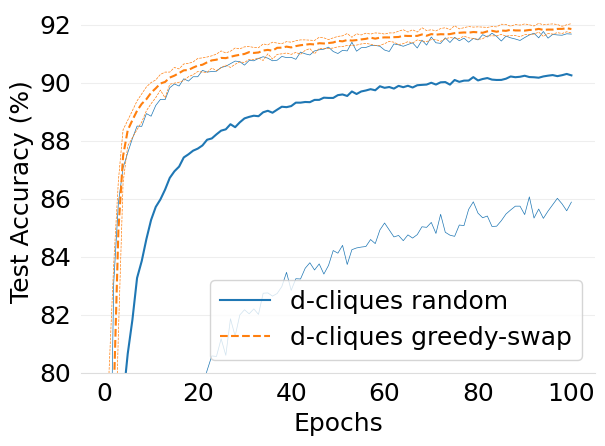
- mlsys2022style/figures/convergence-speed-mnist-dc-random-vs-dc-gs-2-shards-per-node.png 0 additions, 0 deletions...ence-speed-mnist-dc-random-vs-dc-gs-2-shards-per-node.png
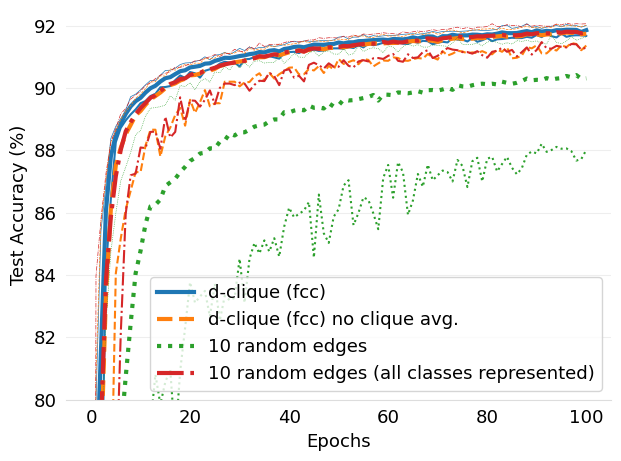
- mlsys2022style/figures/d-cliques-cifar10-linear-comparison-to-non-clustered-topologies.png 0 additions, 0 deletions...cifar10-linear-comparison-to-non-clustered-topologies.png
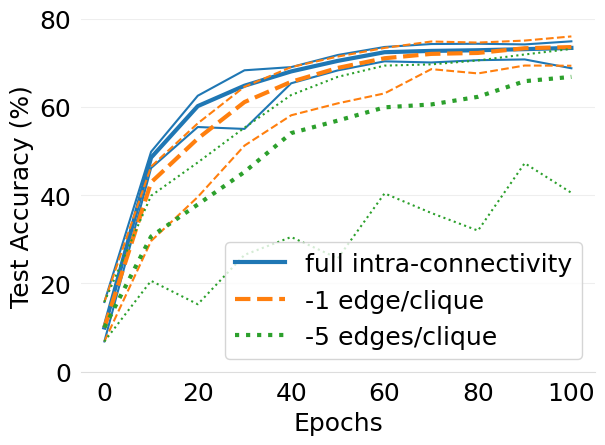
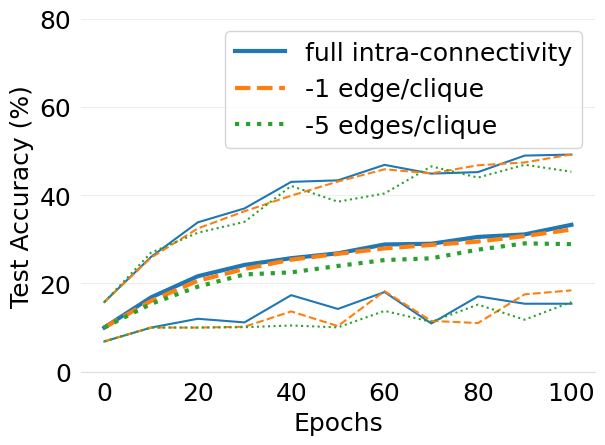
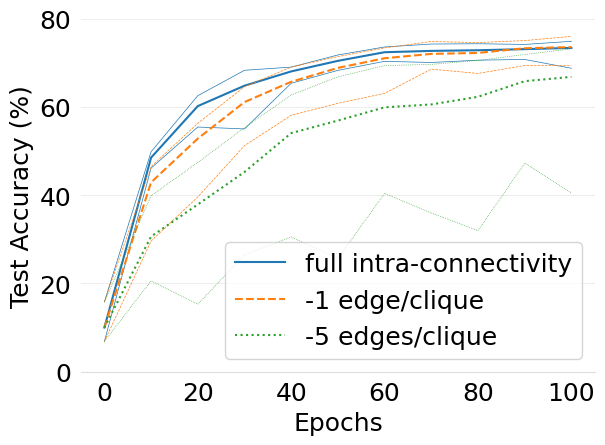
- mlsys2022style/figures/d-cliques-cifar10-w-clique-avg-impact-of-edge-removal.png 0 additions, 0 deletions...d-cliques-cifar10-w-clique-avg-impact-of-edge-removal.png
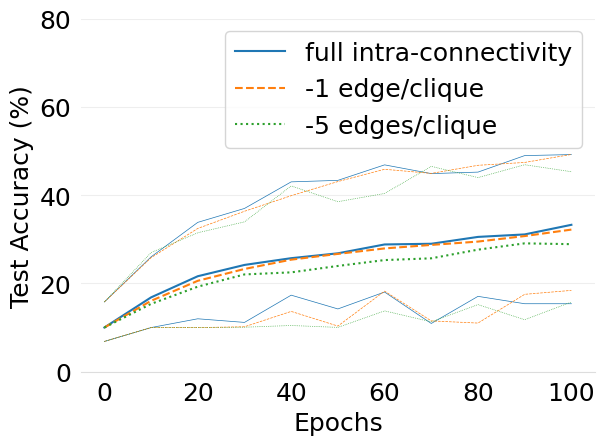
- mlsys2022style/figures/d-cliques-cifar10-wo-clique-avg-impact-of-edge-removal.png 0 additions, 0 deletions...-cliques-cifar10-wo-clique-avg-impact-of-edge-removal.png
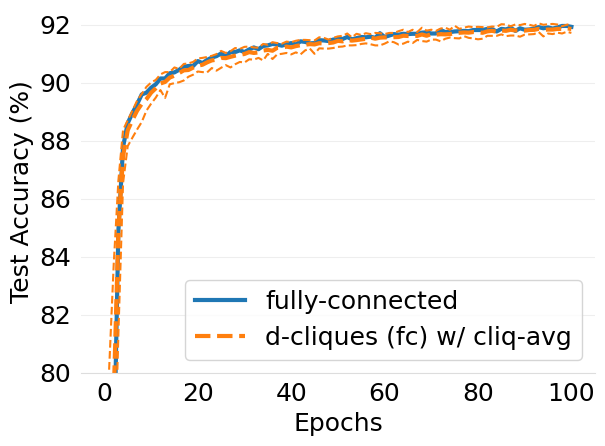
- mlsys2022style/figures/d-cliques-mnist-linear-comparison-to-non-clustered-topologies.png 0 additions, 0 deletions...s-mnist-linear-comparison-to-non-clustered-topologies.png
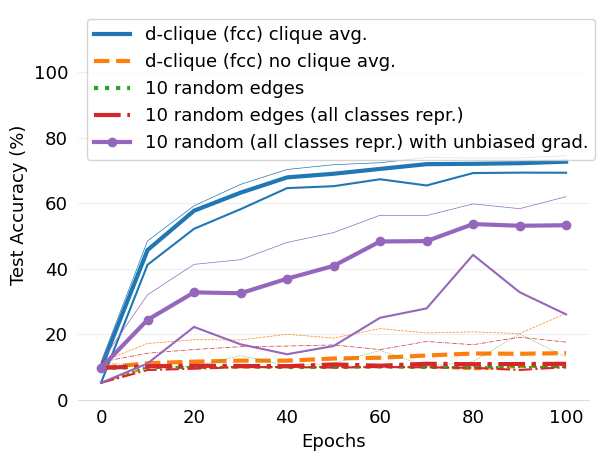
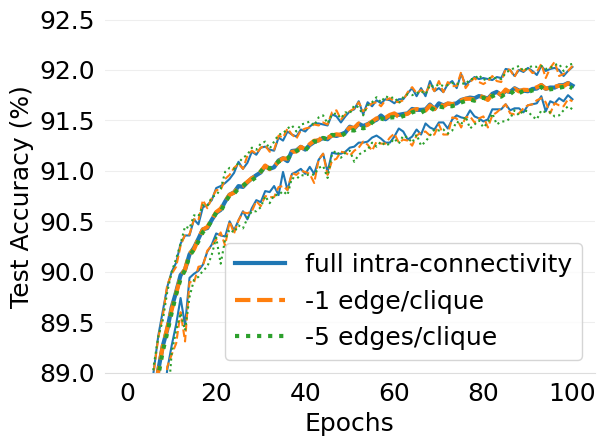
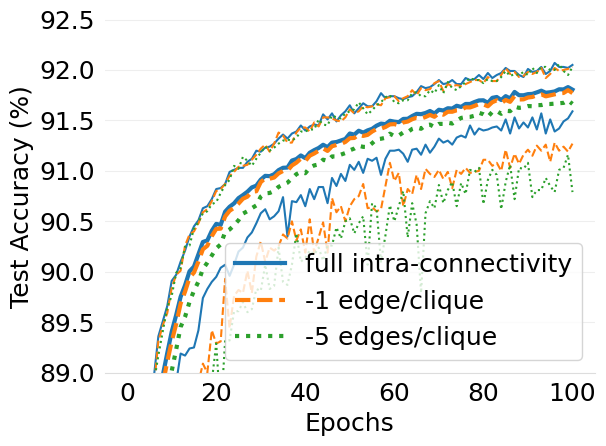
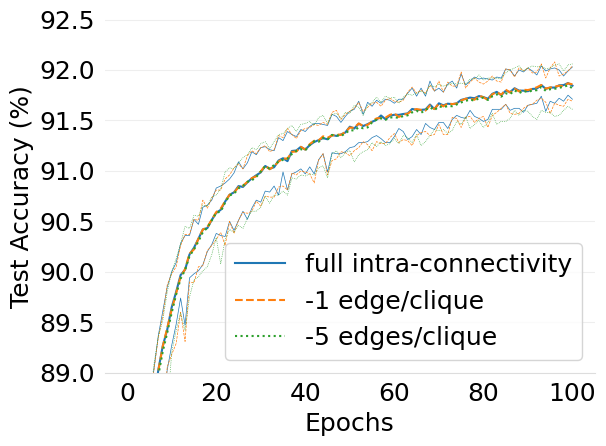
- mlsys2022style/figures/d-cliques-mnist-w-clique-avg-impact-of-edge-removal.png 0 additions, 0 deletions...s/d-cliques-mnist-w-clique-avg-impact-of-edge-removal.png
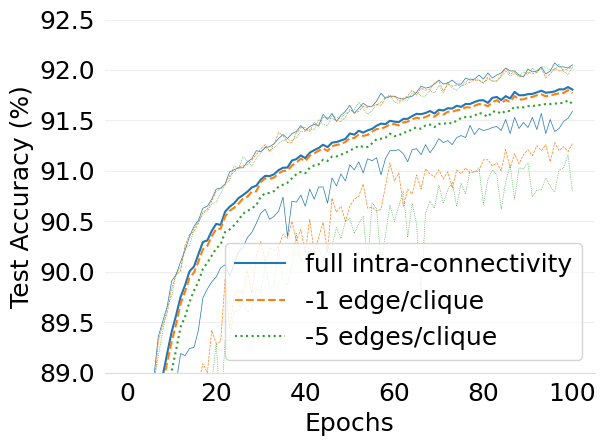
- mlsys2022style/figures/d-cliques-mnist-wo-clique-avg-impact-of-edge-removal.png 0 additions, 0 deletions.../d-cliques-mnist-wo-clique-avg-impact-of-edge-removal.png
| W: | H:
| W: | H:


| W: | H:
| W: | H:


| W: | H:
| W: | H:


| W: | H:
| W: | H:


| W: | H:
| W: | H:


| W: | H:
| W: | H:


| W: | H:
| W: | H:


| W: | H:
| W: | H:


| W: | H:
| W: | H:


| W: | H:
| W: | H:


72.3 KiB
| W: | H:
| W: | H:


| W: | H:
| W: | H:


90.1 KiB
| W: | H:
| W: | H:


| W: | H:
| W: | H: