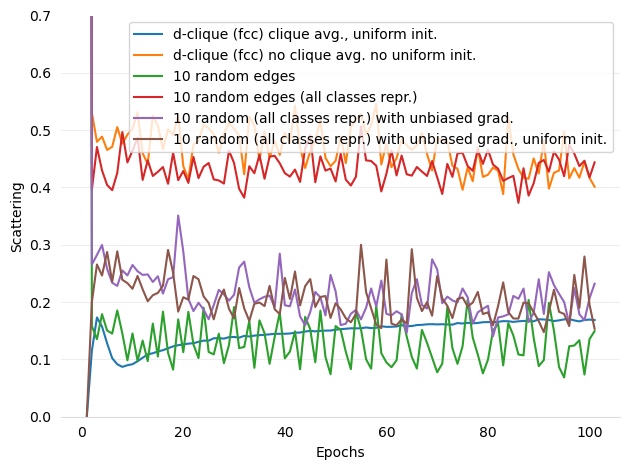
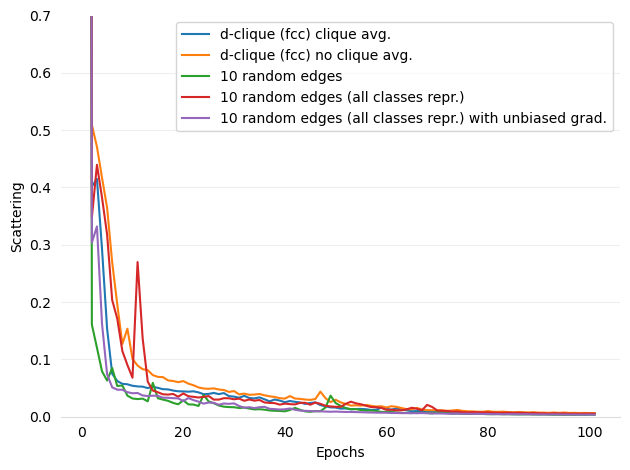
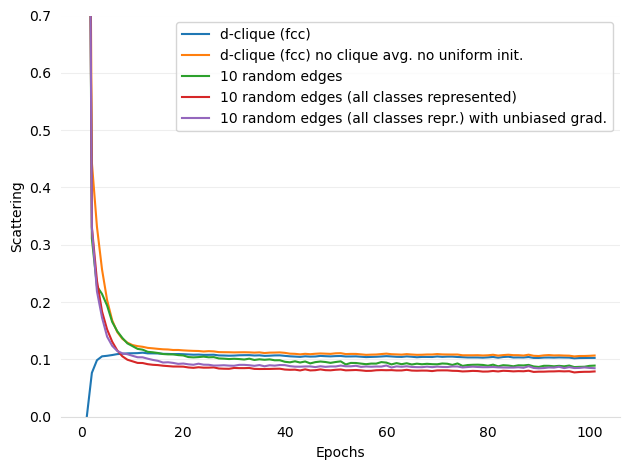
Added figures and ref for Scattering
Showing
- figures/d-cliques-cifar10-linear-comparison-to-non-clustered-topologies-scattering.png 0 additions, 0 deletions...ear-comparison-to-non-clustered-topologies-scattering.png
- figures/d-cliques-mnist-lenet-comparison-to-non-clustered-topologies-scattering.png 0 additions, 0 deletions...net-comparison-to-non-clustered-topologies-scattering.png
- figures/d-cliques-mnist-linear-comparison-to-non-clustered-topologies-scattering.png 0 additions, 0 deletions...ear-comparison-to-non-clustered-topologies-scattering.png
- main.bib 9 additions, 0 deletionsmain.bib
- main.tex 42 additions, 6 deletionsmain.tex
113 KiB
49.1 KiB
46.3 KiB