fix conflict
No related branches found
No related tags found
Showing
- mlsys2022style/appendix.tex 54 additions, 0 deletionsmlsys2022style/appendix.tex
- mlsys2022style/d-cliques.tex 7 additions, 1 deletionmlsys2022style/d-cliques.tex
- mlsys2022style/exp.tex 163 additions, 144 deletionsmlsys2022style/exp.tex
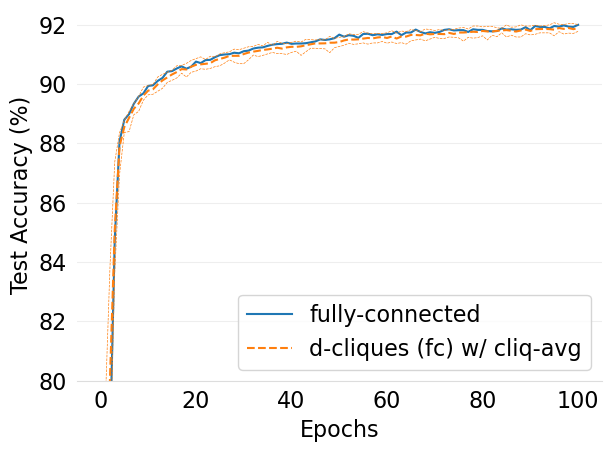
- mlsys2022style/figures/convergence-speed-cifar10-dc-fc-vs-fc-1-class-per-node.png 0 additions, 0 deletions...onvergence-speed-cifar10-dc-fc-vs-fc-1-class-per-node.png
- mlsys2022style/figures/convergence-speed-cifar10-dc-fc-vs-fc-2-shards-per-node.png 0 additions, 0 deletions...nvergence-speed-cifar10-dc-fc-vs-fc-2-shards-per-node.png
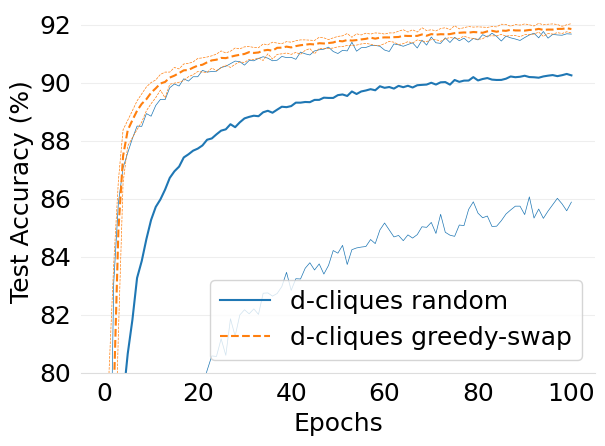
- mlsys2022style/figures/convergence-speed-cifar10-dc-random-vs-dc-gs-2-shards-per-node.png 0 additions, 0 deletions...ce-speed-cifar10-dc-random-vs-dc-gs-2-shards-per-node.png
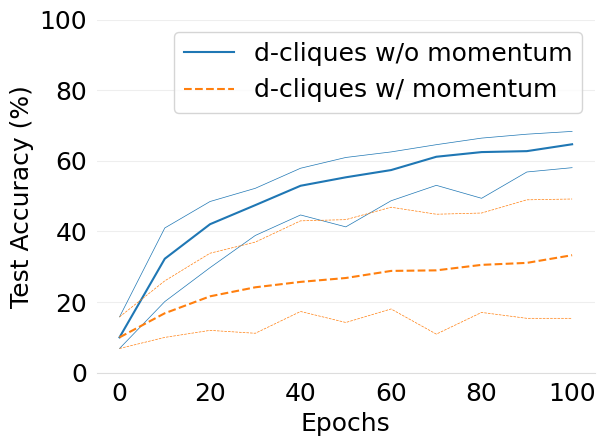
- mlsys2022style/figures/convergence-speed-cifar10-w-c-avg-no-mom-vs-mom-2-shards-per-node.png 0 additions, 0 deletions...speed-cifar10-w-c-avg-no-mom-vs-mom-2-shards-per-node.png
- mlsys2022style/figures/convergence-speed-cifar10-wo-c-avg-no-mom-vs-mom-2-shards-per-node.png 0 additions, 0 deletions...peed-cifar10-wo-c-avg-no-mom-vs-mom-2-shards-per-node.png
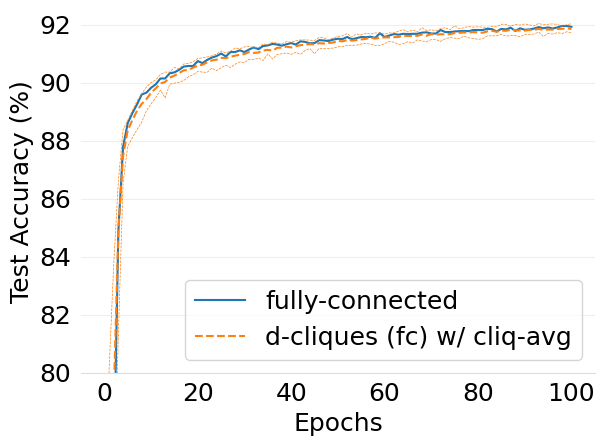
- mlsys2022style/figures/convergence-speed-mnist-dc-fc-vs-fc-1-class-per-node.png 0 additions, 0 deletions.../convergence-speed-mnist-dc-fc-vs-fc-1-class-per-node.png
- mlsys2022style/figures/convergence-speed-mnist-dc-fc-vs-fc-2-shards-per-node.png 0 additions, 0 deletions...convergence-speed-mnist-dc-fc-vs-fc-2-shards-per-node.png
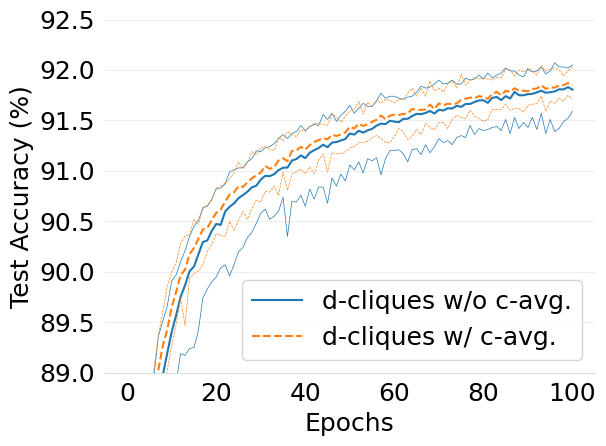
- mlsys2022style/figures/convergence-speed-mnist-dc-no-c-avg-vs-c-avg-2-shards-per-node.png 0 additions, 0 deletions...ce-speed-mnist-dc-no-c-avg-vs-c-avg-2-shards-per-node.png
- mlsys2022style/figures/convergence-speed-mnist-dc-random-vs-dc-gs-2-shards-per-node.png 0 additions, 0 deletions...ence-speed-mnist-dc-random-vs-dc-gs-2-shards-per-node.png
- mlsys2022style/figures/convergence-speed-mnist-dc-rm-1-edge-vs-full.png 0 additions, 0 deletions.../figures/convergence-speed-mnist-dc-rm-1-edge-vs-full.png
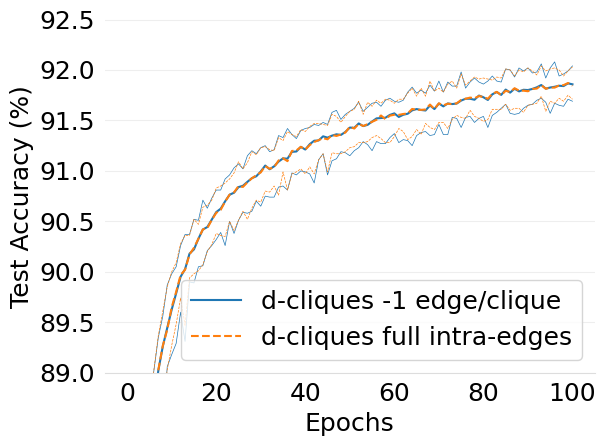
- mlsys2022style/figures/d-cliques-ideal-w-clique-avg-impact-of-edge-removal.png 0 additions, 0 deletions...s/d-cliques-ideal-w-clique-avg-impact-of-edge-removal.png
- mlsys2022style/figures/d-cliques-ideal-wo-clique-avg-impact-of-edge-removal.png 0 additions, 0 deletions.../d-cliques-ideal-wo-clique-avg-impact-of-edge-removal.png
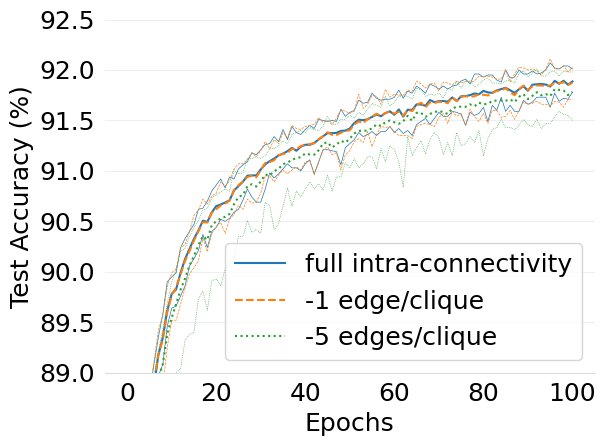
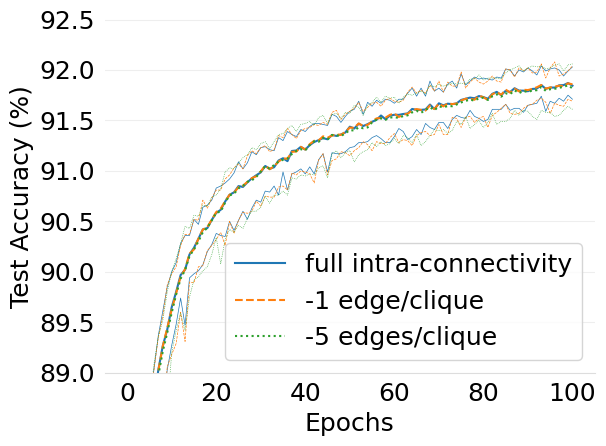
- mlsys2022style/figures/d-cliques-w-clique-avg-impact-of-edge-removal.png 0 additions, 0 deletions...figures/d-cliques-w-clique-avg-impact-of-edge-removal.png
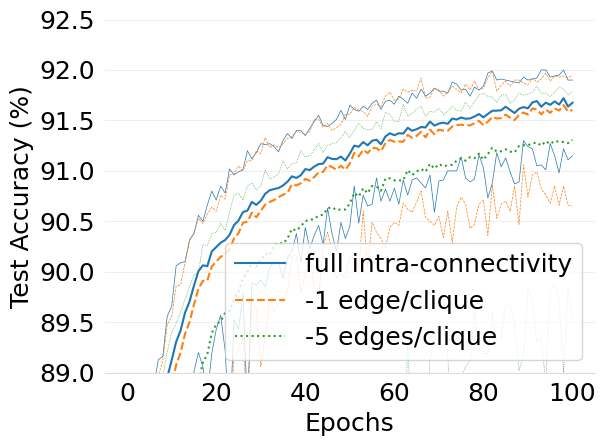
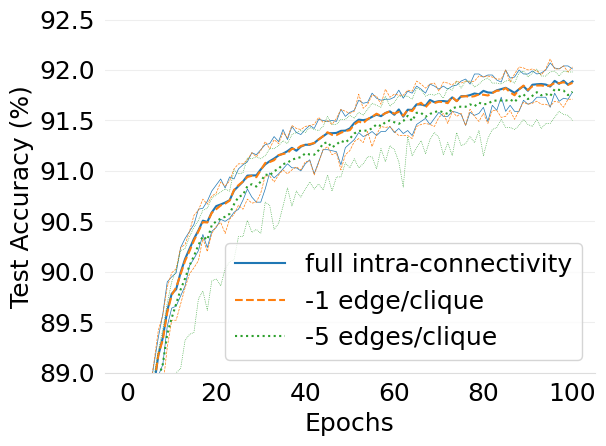
- mlsys2022style/figures/d-cliques-wo-clique-avg-impact-of-edge-removal.png 0 additions, 0 deletions...igures/d-cliques-wo-clique-avg-impact-of-edge-removal.png
- results-v2/dc-greedy-swap-rm-edges-cifar10-2-shards-eq-classes-fc.sh 5 additions, 2 deletions...dc-greedy-swap-rm-edges-cifar10-2-shards-eq-classes-fc.sh
- results-v2/dc-ideal-rm-edges-cifar10-max-local-skew-fc.sh 4 additions, 1 deletionresults-v2/dc-ideal-rm-edges-cifar10-max-local-skew-fc.sh
This diff is collapsed.
45.3 KiB
43.9 KiB
44.6 KiB
46.6 KiB
45.3 KiB
42.7 KiB
44.6 KiB
62 KiB
56.5 KiB
60.7 KiB
78.3 KiB
90.5 KiB
72.4 KiB
78.3 KiB